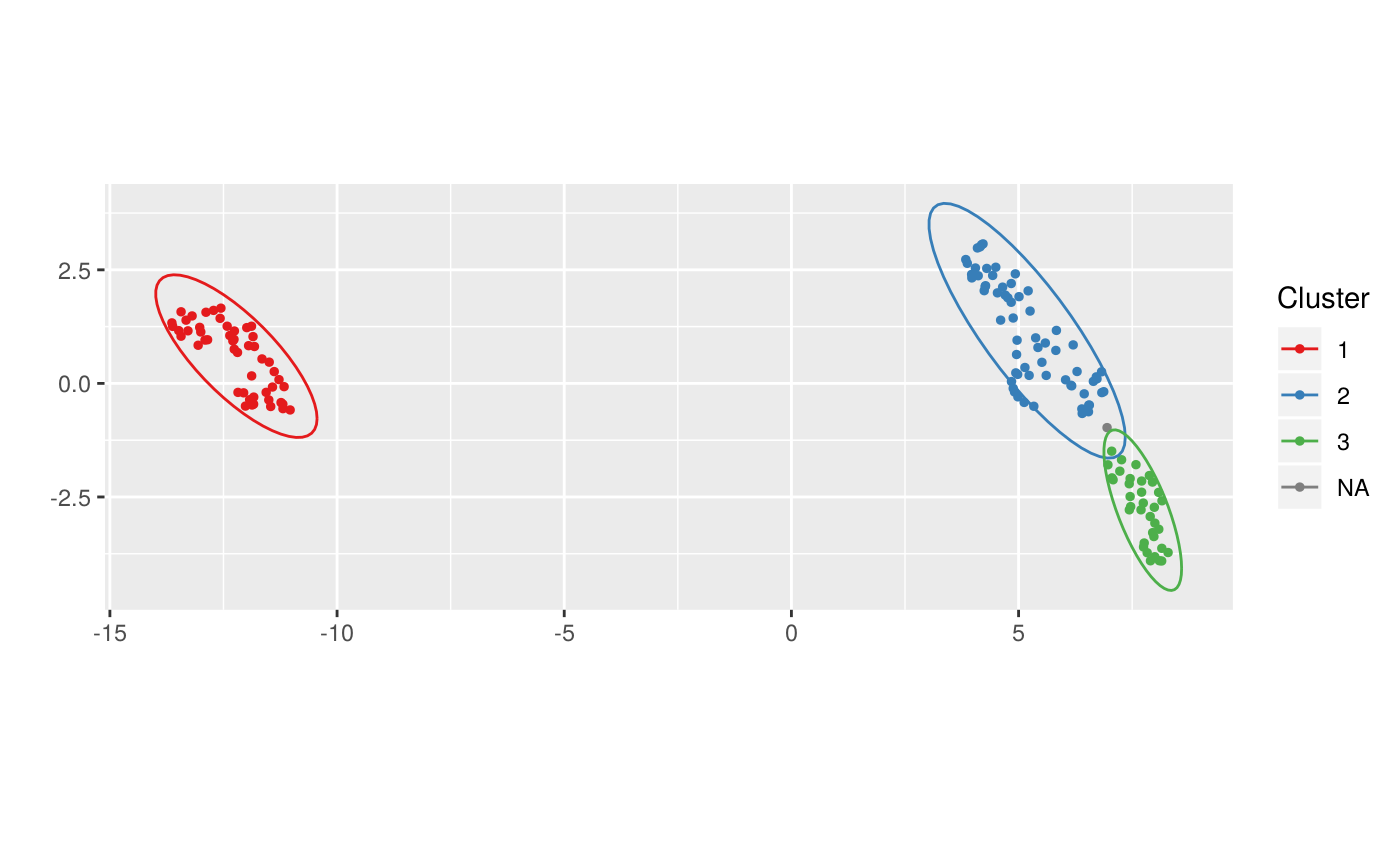
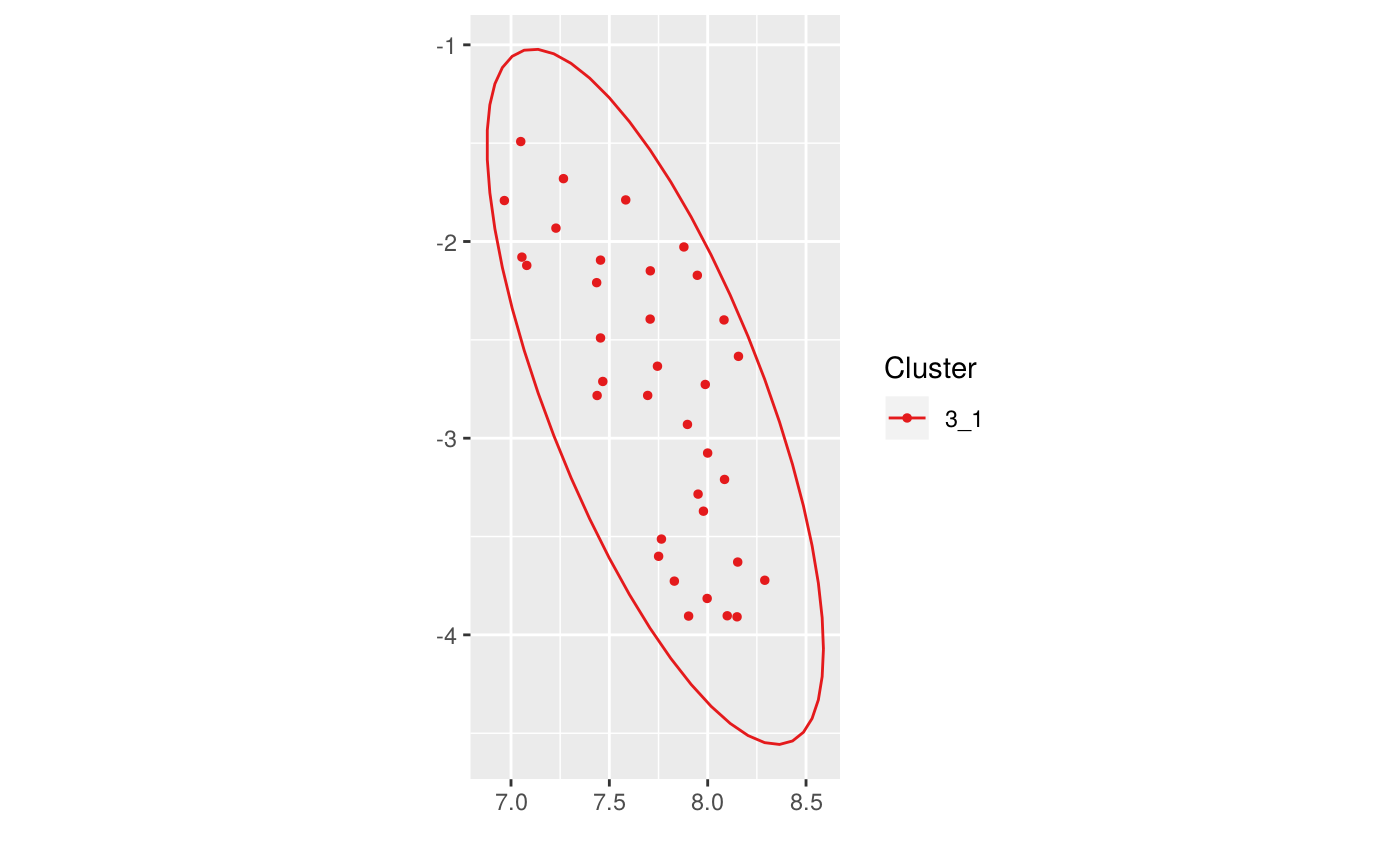
This function runs DBSCAN with the specified arguments to compute new clusters on a umapscan object. Only points which don't have an already validated cluster are taken into account.
compute_clusters( us, parent = "", noise_only = FALSE, eps, minPts, graph = TRUE, alpha = 1, ellipses = TRUE )
Arguments
| us | umapscan object |
|---|---|
| parent | name of the parent cluster |
| noise_only | only compute clusters for current 'Noise' points |
| eps |
|
| minPts |
|
| graph | if TRUE, display a plot of the computed clusters |
| alpha | point transparency for clusters plot |
| ellipses | if TRUE, plot confidence ellipses around clusters |
Value
Returns an updated umapscan object, and optionally displays a clusters plot.
See also
new_umapscan(), describe_clusters(), get_cluster_data(), rename_cluster(),
plot_clusters(), get_clusters_membership(), remove_cluster()
Examples
library(dplyr)#> #>#> #> #>#> #> #>iris_num <- iris %>% select_if(is.numeric) us <- new_umapscan(iris_num, n_neighbors = 25, min_dist = 0.1, seed = 1337) us <- compute_clusters(us, minPts = 3, eps = 0.5)compute_clusters(us, minPts = 3, eps = 0.45, alpha = 1, parent = "3")#> #> Call: new_umapscan(d = iris_num, n_neighbors = 25, min_dist = 0.1, #> seed = 1337) #> #> UMAP embeddings of a 150x4 data frame #> #> Clusters : #> #> levelName #> 1 #> 2 ¦--1 #> 3 ¦--2 #> 4 ¦--3 #> 5 ¦ °--3_1 #> 6 °--<Noise>